MAPL-Molecular Assembly Patterning by Lift-off for Biological Applications
Project
Chemical patterning is of great importance for biological applications such as biosensors as well as for fundamental cell studies (cell-surface interactions).
MAPL is a novel patterning technique where a specific ligand (or multiple ligand types) can be immobilized with a controlled surface density inside a defined area. The role of the ligands is to capture molecules from the environment (proteins in solution; cell transmembrane proteins such as integrins; DNA/RNA). These specific types of interactions should only occur in the designated patterns and not elsewhere. To avoid non-specific interactions the surface between the patterns (background) is passivated. Poly(ethylene glycol) (PEG) chemistry is efficiently used to repel proteins from the surface.
Goal
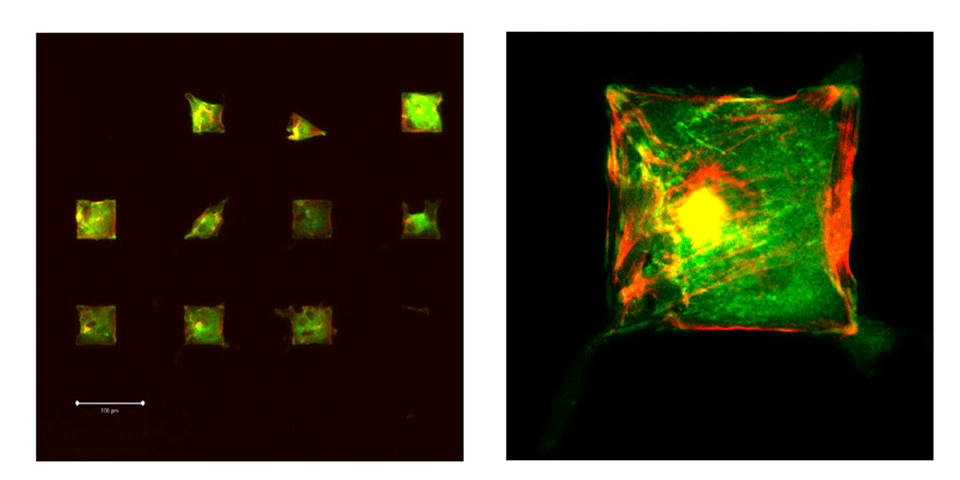
Produce patterned surfaces for various purposes (biosensors and cell patterning) with a well defined chemistry. The patterned surfaces should fulfill the role of a new tool to investigated cell-surface interactions. The aim is to immobilize single cell on individual cell-adhesive patches with controlled geometry and chemistry. The identical cell microenvironment will allow gene expression analysis on a number of cells that have a similar phenotype.
Materials and Method
The MAPL process is carried out in two main steps:
- Transferring the pattern into a resist; for structures down to 1 micrometer (micro-MAPL), standard photolithography is used, while for sub-micrometer features (nano-MAPL) nano hot-embossing is applied.
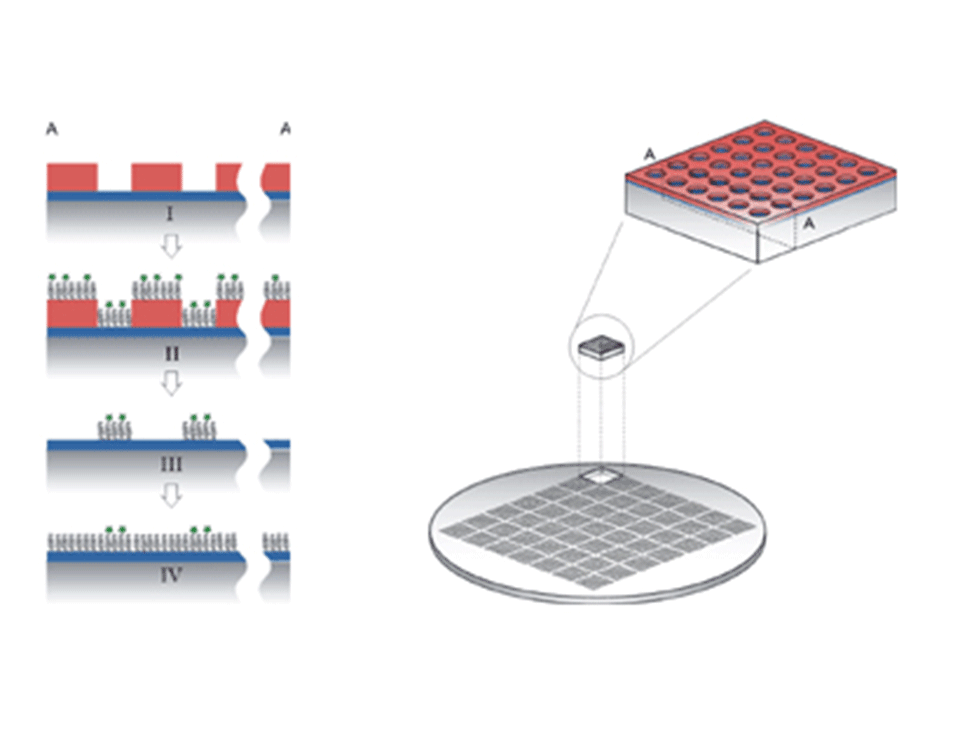
- The pre-patterned surfaces are chemically modified to transfer the resist pattern into a bio-chemical pattern (Figure 1)
A glass or silicon wafer is coated with a 12 nm layer of Nb2O5 or TiO2. The wafer is then spun coated with a layer of photoresist. The resist is illuminated with UV light through a mask with the desired patterns and then developed.
Step I: the chip with a contrast between the oxide and the resist is cleaned
Step II: the sample is dipped into an aqueous solution of poly(L-lysine) grafted poly(ethylene glycol) (PLL-g-PEG) on which a controlled amount of PEG chains are functionalized (e.g. biotin or RGD peptides).
Step III: the resist is lifted-off in an organic solvent without loosing or harming the polymer layer.
Step IV: the background is rendered protein resistant through the adsorption of non-functionalized PLL-g-PEG. The result is a (bio-)chemical pattern with a controlled density of ligands.
Nano-MAPL
Structures smaller than one micrometer are difficult to achieve with classical lithographic techniques. To overcome the size limitation we apply a nano hot-embossing (figure 2) process (PSI, Villigen, Switzerland). PMMA is used in place of the traditional photoresist; the melted PMMA layer is embossed with a stamp of nano-structures. To obtain the PMMA/oxide contrast the embossed sample is etched with O2 plasma.
Results
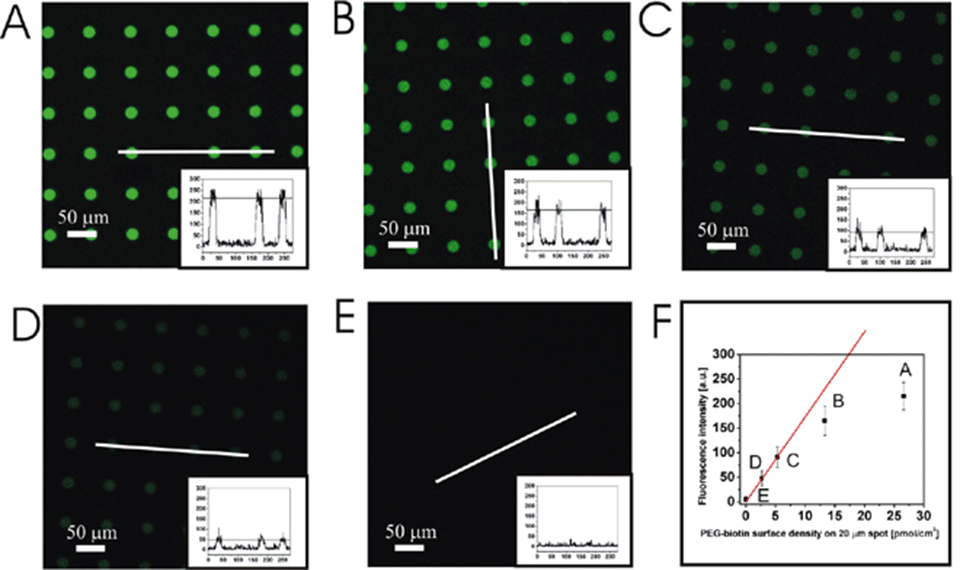
By mixing the functionalized PLL-g-PEG with non-functionalized polymer the surface density can be decreased in a controlled manner. Figure 2 shows the fluorescent intensity of the streptavidin (alexa 488 labeled) specifically bound to the biotin functionalized PLL-g-PEG.